bioparticle
a PFLOTRAN reaction sandbox for particle transport
Kinetic attachment/detachment continuum-based model for nanoparticles transport in porous media
What is this experiment?
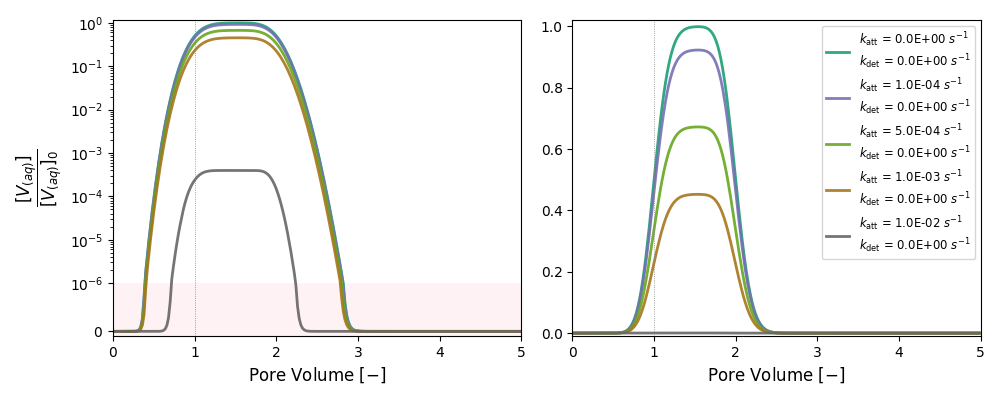
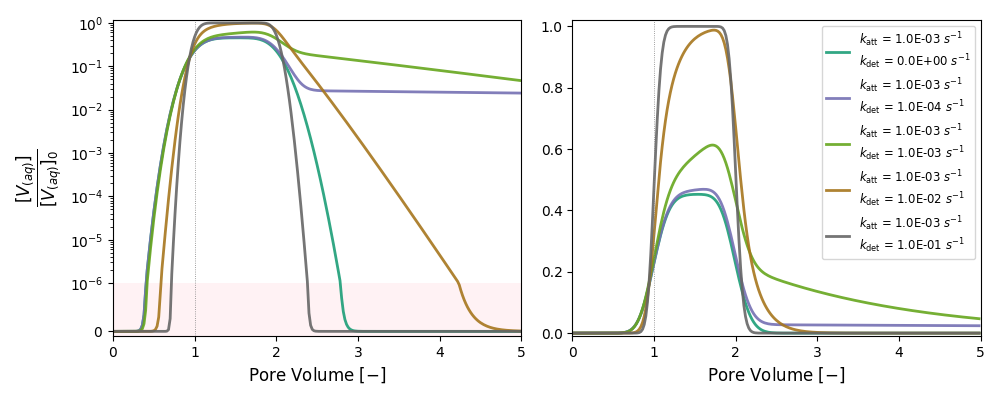
It simulates breakthrough curves of nanoparticles for attachment and detachment rates that span several orders of magnitude.
What does the code do?
runMultipleCases.py reads a CSV file with a list of cases to run in PFLOTRAN, allowing the variation of parameters that are pointed out in the template .IN file as follows:
REACTION_SANDBOX
BIOPARTICLE
RATE_ATTACHMENT <katt> 1/s
RATE_DETACHMENT <kdet> 1/s
/
/
the tags <katt> and <kdet> are replaced for the list of values indicated in the csv-file, in the column with the same header.
How to run this test?
$ python3 runMultipleCases.py [CSV_PARAMETERS] [TEMPLATE_FILE] -run
Where:
[CASES.CSV]: path to csv file with the list of parameters and the corresponding tags[TEMPLATE.IN]: input file template for PFLOTRAN and the corresponding tags
References for this experiment:
-
Babakhani, P., Bridge, J., Doong, R., & Phenrat, T. (2017). Continuum-based models and concepts for the transport of nanoparticles in saturated porous media: A state-of-the-science review. Advances in Colloid and Interface Science, 246, 75–104.
-
Babakhani, P., Fagerlund, F., Shamsai, A., Lowry, G. V., & Phenrat, T. (2015). Modified MODFLOW-based model for simulating the agglomeration and transport of polymer-modified Fe0 nanoparticles in saturated porous media. Environmental Science and Pollution Research, 25(8), 7180–7199.
Description
An injection of nanoparticles at a given concentration is set at the inlet of a column experiment. After some time, the nanoparticle injection is stopped and only clean water keeps runing through the column.
| Column parameters | Value | Unit | |
|---|---|---|---|
| Lenght | L | 25.5 | cm |
| Diameter | Ø | 1.27 | cm |
| Grain size | d50 | 0.300 | mm |
| Porosity | φ | 0.33 | - |
| Long. Dispersivity | αL | 0.015 | cm |
| Particle parameters | Value | Unit | |
|---|---|---|---|
| Size | dp | ?? | nm |
| Initial concentration | C0 | 1.0 × 10-5 | mol/L |
List of parameters
| Set |
Case | katt | kdet |
|---|---|---|---|
| Only Attachment | 1A | 0 | 0 |
| 2A | 1.0 × 10-4 | 0 | |
| 3A | 5.0 × 10-4 | 0 | |
| 4A | 1.0 × 10-3 | 0 | |
| 5A | 1.0 × 10-2 | 0 | |
| Vary Detachment | 1B | 1.0 × 10-3 | 0 |
| 2B | 1.0 × 10-3 | 1.0 × 10-4 | |
| 3B | 1.0 × 10-3 | 1.0 × 10-3 | |
| 4B | 1.0 × 10-3 | 1.0 × 10-2 | |
| 5B | 1.0 × 10-3 | 1.0 × 10-1 |
k and λ units in [s-1]
PFLOTRAN Simulation